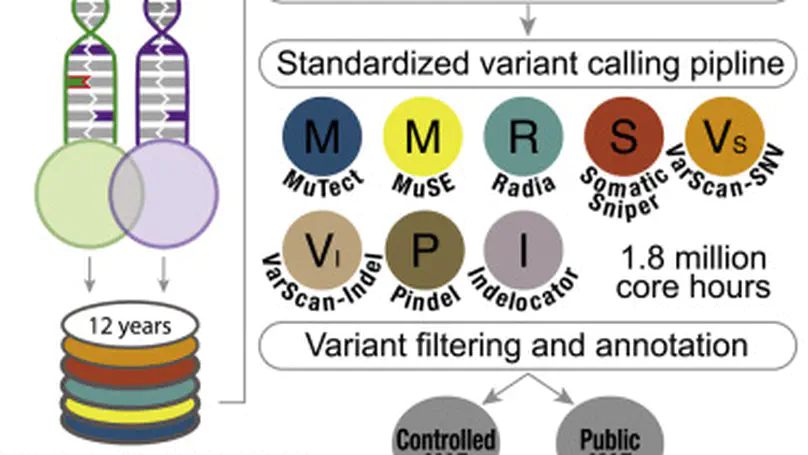
Summary Molecular subtypes, such as defined by The Cancer Genome Atlas (TCGA), delineate a cancer’s underlying biology, bringing hope to inform a patient’s prognosis and treatment plan. However, most approaches used in the discovery of subtypes are not suitable for assigning subtype labels to new cancer specimens from other studies or clinical trials. Here, we address this barrier by applying five different machine learning approaches to multi-omic data from 8,791 TCGA tumor samples comprising 106 subtypes from 26 different cancer cohorts to build models based upon small numbers of features that can classify new samples into previously defined TCGA molecular subtypes—a step toward molecular subtype application in the clinic. We validate select classifiers using external datasets. Predictive performance and classifier-selected features yield insight into the different machine-learning approaches and genomic data platforms. For each cancer and data type we provide containerized versions of the top-performing models as a public resource.