MC3
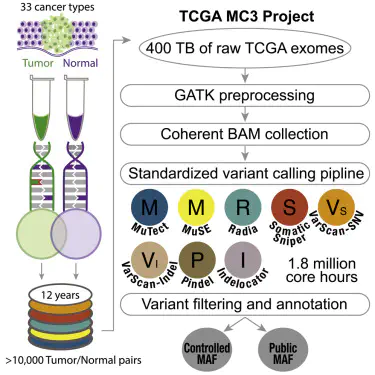
The TCGA project combined DNA sequence data from three centers generated at Broad, Baylor, and Washington University. Mutation calling was performed independently at one or more of these centers for each of the tumor specific marker papers. The methods used for mutation calling varied as the project evolved over the years. The TCGA PanCancer Atlas MC3 set is a re-calling of uniform files to remove batch effects and enable pancancer analysis. We recognize the advantage to doing a uniform analysis where all the TCGA exome data was called on a standardized set of mutation callers. Further, we recognize the value of “containerizing” the procedures used, so that the methods are transparent and reproducible and sharable with the community. As detailed on this wikipage, the TCGA MC3dataset represents a uniform set of mutation calls. The BAM files used underwent a standardized local re-alignment to hg19 (Genome Reference Consortium GRCh37), six calling algorithms were applied, and a number of automated filters were applied. We strongly recommend that users review the information provided on caveats, data processing, filters, best practices, and acknowledgements before using this file.